Characterization of proteins and post-translational modifications by liquid-chromatography mass spectrometry
Over the past three decades, mass spectrometry and the combination of liquid-chromatography with mass spectrometry (LC-MS) became one of the most powerful techniques for the identification and characterization of proteins and post-translational modifications. While originally triple quadrupole mass spectrometers have been used, these have later usually been replaced first by ion traps and later by either QTOF or Orbitrap technology based systems because of their higher resolution, speed and mass accuracy.
Mass spectrometry and proteomics became a dominant area of research which deals with high-throughput protein identification, analysis of isoforms and modifications, alternative gene products, disease origins, etc.
Also, many aspects of critical quality attributes (CQA) of biologics are nowadays investigated by LC-MS and protein mass spectrometry.
How can mass spectrometry help in the analysis of proteins?
Proteins consist out of a usually linear sequence of 22 amino acids (actually more if some specific variants are counted as well). Most of these amino acids have distinct elemental compositions and molecular masses.
When a protein is synthesized, these amino acids assemble into a chain where the mass is determined by the masses of the amino acids from which it is formed. Comparing the measured mass of the intact protein can tell us whether there is a difference between the mass we calculated from the amino acid composition or not. In the first case either the expected compostion is wrong or a so-called post-translational modification introduced a change after the synthesis of the protein chain.
The analysis of the intact protein by mass spectrometry can provide us with a first information about the integrity of the protein.
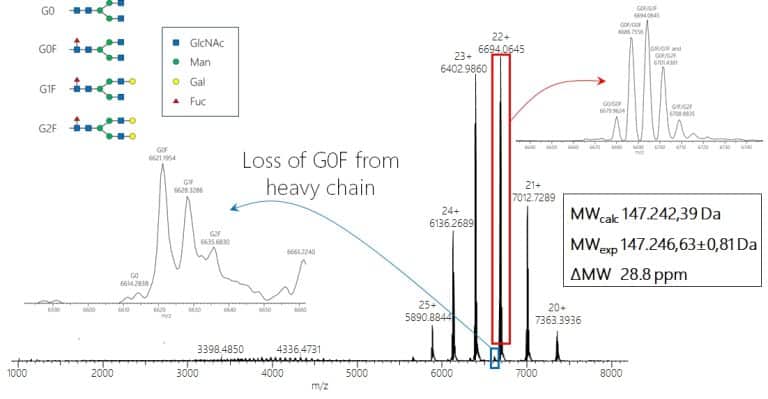
Some post-translational modifications can also be heterogeneous, such as glycosylations. These modifcations then also introduce a heterogeneity into the intact modified protein so that multiple masses for one protein may appear in the mass spectrum. A typical example for this are monoclonal antibodies which carry (at least) a conserved glycosylation at both heavy chains.
While for biologics the protein sequence is usually known, for proteins from biological samples it might not be known. This is the area of classical proteomics analysis using protein identification by by mass spectrometry.
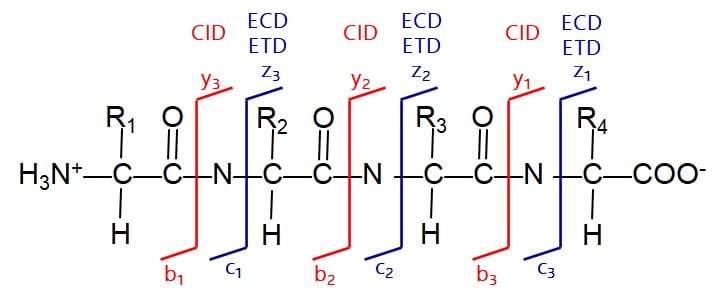
Here the intact protein is commonly digested using specific enzymes (most commonly Trypsin), which cleave the protein chain at specific residues int smaller, more handy peptides. These peptides are then separated by liquid chromatography and fragmented in the mass spectrometer. Under collision-induced dissociation conditions, the fragmentation of the peptide chain usually always occurs within the amide bond between the amino acid residues. As a result we obtain a spectrum with specific peaks showing a spacing which corresponds to the amino acids forming the peptide on our triple quadrupole-, QTOF- or Orbitrap-based mass spectrometer. With a set of peptides and the amino acid sequence derived from their MS/MS spectra we can now run a database search to identify the protein based on a sequence originally obtained e.g. from genetic sequencing and being stored in databases.
How can MS Vision help you with that?
MS Vision is specialized on liquid chromatography-mass spectrometry (LC-MS) technology. We offer dedicated services around this technology like
- preventive maintenance and repairs of LC-MS systems and mass spectrometers
- providing used and refurbished instrumentation (mass spectrometry systems from Waters, Sciex and ThermoFisher as well as LC systems from Waters, Sciex, ThermoFisher, Agilent and Shimadzu) including shipment, installation, warranty and after sales support
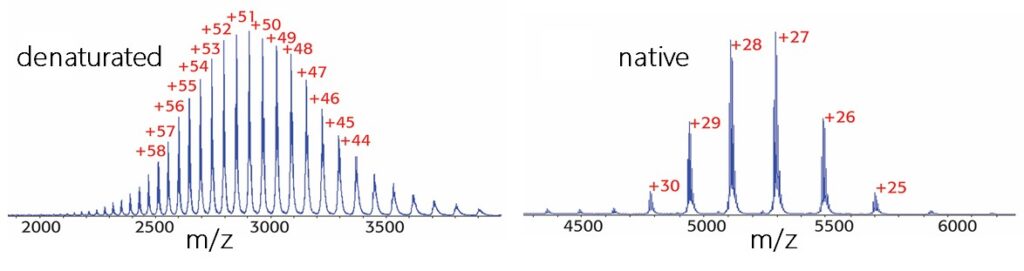
- modification of Waters Synapt and ThermoFisher Q Exactive mass spectrometers for the analysis of intact proteins under native ESI conditions and the characterization of non-covalent protein complexes or AAV particles.
