We regret to inform you that after acquisition of our collaborator e-MSion, Inc. by Agilent Technologies in March, our distribution agreement has recently been terminated by Agilent with 6 months notice. That means we are currently working on finalizing current projects, yet Agilent Technologies decided to stop production and distribution of ECD upgrades for Waters and ThermoFisher instruments in the future.
Customers with cell upgrades on Waters instruments will be able to purchase filaments directly from Agilent after the distribution agreement has ended, the filaments for Waters and Agilent cells are identical. Customers with ThermoFisher cell upgrades will be able to purchase any amount of filaments in the next 6 months (we recommend to place orders within the next four months to ensure proper handling and processing within six months) to use them in the future. In parallel we are trying to find alternative sources for the filaments for these customers.

Ion mobility and ECD
Top-down mass spectrometry
Deamidation

De-novo sequencing
e-MSion‘s ExD cell provides you with a range of extended capabilities for running your experiments. The tunable electron energy (0.3-100 eV) allows to analyze compounds from small lipids up to multimeric megadalton complexes.
Typical applications are:
- top-down analysis of proteins, biologics and non-covalent complexes
- analysis of labile or difficult post-translational modifications (PTM’s) such as phosphorylations (including those on Lys, Arg and His residues), sulfatations, deamidation (Asp/isoAsp differentiation) or γ-Carboxyglutamate
- Hydrogen/deuterium exchange (HDX) for protein structure characterization
- collisional induced unfolding for protein structure analysis
- de-novo sequencing (Leu/Ile differentiation)
- glycan intraring cleavage for connectivity analysis
- lipid fragmentation
Some of the applications require the usage of IMS in combination with ECD and are thus exclusively available on Waters Synapt technology.
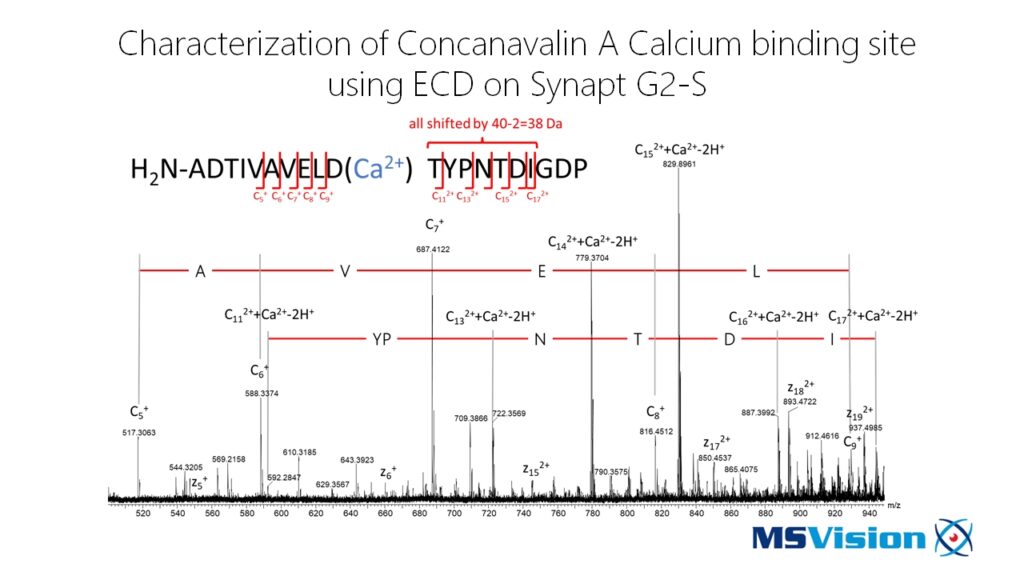
Enhanced Structural Information
For proteins and peptides, ECD has the ability to indiscriminately cleave the backbone N-Cα bond, while leaving the more labile side-chain modifications intact. Combining ECD and CID spectra, can potentially provide more complete data for de novo sequencing. Because post translational modifications such as phosphorylation, carboxylation, glycosylation, and sulfation are less easily lost in ECD than in CID, ECD assignments of their sequence positions are far more specific. This is particularly useful when studying for example protein interactions or conducting structural biology experiments. Combining this with ion mobility adds even more potentially useful information.
When aiming for structural information by H/D exchange, fragmentation by ECD – in contrast to CID – is not causing hydrogen scrambling. This makes the analysis of the obtained data much more easy. Furthermore, ECD is able to efficiently fragment bigger peptides than CID giving you access to areas with only a few enzymatic cleavage sites, such as e.g. transmembrane domains, for H/D exchange analysis.
Don’t I need expensive instruments for it though?
In 2015 e-MSion (https://www.agilent.com/en/product/liquid-chromatography-mass-spectrometry-lc-ms/e-msion) was founded to make ECD available to the wider MS community as a retrofittable device on non-FT spectrometers. At MS Vision we have many years of experience of making modifications to existing hardware to improve its. We are happy to join forces with e-MSion to bring this technology to owners of Waters Synapt and ThermoFisher QExactive (except of QExactive UHMR) instruments as an upgrade option.
What does it give me that I can’t already get?
The ECD cell produces ECD fragmentation similar to FTICR-ECD, but on workhorse mass spectrometers like Q-TOFs and Orbitrap QEs. ECD tends to produce mainly c and z type ions and as a result the spectra produced tend to be less congested thus making interpretation and top down sequencing simpler. The lower energies allow for the retention of PTMs and thereby allow for the identification of the site of PTMs such as phosphorylation and glycosylation etc especially when combined with Native MS (https://dx.doi.org/10.1021/acs.analchem.9b04763). The ECD cell enables single-residue-resolution HDX-MS without hydrogen scrambling and disulfide bond localization without prior reduction. All of this can be achieved in conjunction with UHPLC, HPLC, CE and IMS thus making the technique compatible with your standard workflows. A list of example data can be found on the e-MSion website https://www.agilent.com/en/product/liquid-chromatography-mass-spectrometry-lc-ms/e-msiontechnology/example-data/.
